DNA Methylation Detection
DNA methylation detection is a detection method that uses a variety of methods to determine the degree of methylation in malignant tumor cells in order to better judge the degree and spread of the lesion. DNA methylation is an important mechanism to regulate gene expression. The state of methylation is not invariable in the development of malignancy. Genome-wide hypomethylation in tumor cells is closely related to disease progression, tumor size and malignancy. Therefore, this test is very important for judging the degree of malignancy of the tumor.
Technical principle
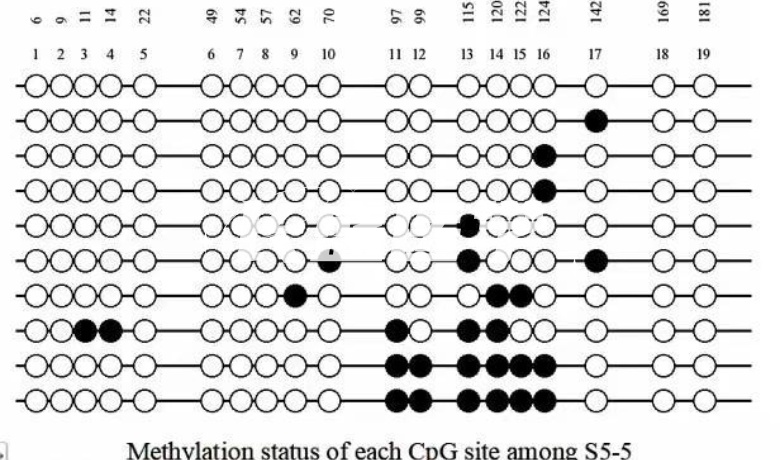
NA methylation is an important mechanism of gene expression regulation, and DNA methylation detection is the use of a variety of methods to determine the level of DNA methylation in tumor cells. In the development of malignant tumors, the degree of methylation is not static, and the degree of hypomethylation in tumor cells is closely related to the deterioration of the disease, tumor volume and the degree of malignancy. Therefore, DNA methylation detection is very important for the evaluation of tumor malignancy. Methylation-specific PCR(MSP) is a specific methylation PCR method proposed by Herman et al. in 1996. The method utilizes sulfite treatment of genomic DNA to convert unmethylated cytosines to uracil, while methylated cytosines are unaffected; primers specific for methylated and unmethylated sequences are then designed for PCR. The MSP amplification product is detected by electrophoresis. If the primer for the methylated DNA chain can amplify the fragment, it indicates that the site is methylated; otherwise, it indicates that the detected site is not methylated. The MSP method has a wide range of applications and can detect whether specific sites are methylated. Bisulfite sequencing PCR(BSP), a bisulfite-modified sequencing method, was proposed by Frommer et al. in 1992. The method has high reliability and high accuracy, and can determine the methylation status of each CpG site in the target fragment. Treatment of genomic DNA with bisulfite converts unmethylated cytosines to uracil, while methylated cytosines are unaffected. BSP primers are then designed for PCR, during which all uracil is converted to thymine. Finally, the PCR product is sequenced to determine whether the CpG site is methylated. In addition, the use of cloning technology can improve the success rate of sequencing, this method is called BSP-cloning sequencing. The BSP method is highly accurate and can accurately detect the percentage of methylation.
Real Experimental Research Hundreds of Detection Experiments 6 Experimental Platforms