SNP
Single nucleotide polymorphism refers to the DNA sequence polymorphism caused by the variation of a single nucleotide. This is the most prevalent type of heritable variation in humans, accounting for approximately over 90% of all known polymorphisms. SNPs (Single Nucleotide Polymorphism) are very common in the human genome, with an average of one in every 300 base pairs, and the total number is estimated to reach 3 million or more. Such dimorphic labeling is caused by a single base transition or transversion, but also by a base insertion or deletion. In addition to variations that may occur within a gene sequence, SNPs may also be present at non-coding sequences outside of a gene.
Technical principle
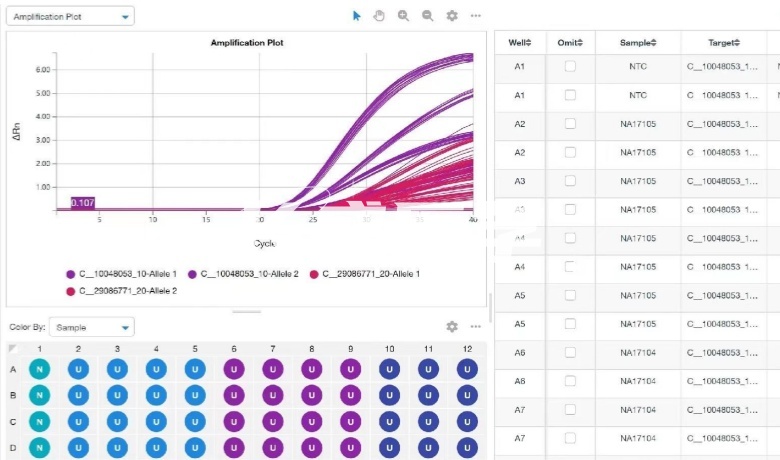
TaqMan probe technology for different SNP sites on the chromosome PCR amplification, and by real-time fluorescent PCR method for detection. The 5 '-end and the 3'-end of this probe are labeled with different fluorescent groups, achieving efficient detection. When the PCR product is present in the solution, the probe and the template can be annealed to produce a structure compatible with the exonuclease-specific substrate, the fluorescent group on the 5 '-end of the probe is cleaved, and the PRET between the probes is destroyed, thereby emitting a fluorescent signal. This technique is commonly used to analyze a small number of SNP sites. High-resolution melting curve (HRM) is an emerging SNP research tool in recent years. It is through the real-time detection of PCR amplification products and double-stranded DNA fluorescent dye in the process of heating, to determine the existence of SNP. Different SNP loci and whether they are heterozygous will affect the peak shape of melting curve, so HRM analysis can accurately analyze different SNP loci and different genotypes. This detection method is not limited to the mutation base site and type, and does not require sequence-specific probes. It only needs high-resolution melting after PCR to quickly obtain the genotype of the sample. This method does not need to design the probe, simple, low cost, accurate results, and can achieve real closed tube operation. SNaPshot technology is a SNP typing technology based on the principle of fluorescent labeling single base extension, also known as small sequencing technology. The technique is aimed at a medium-throughput SNP typing project by extending the primer by one base in a reaction system containing a sequencer, four fluorescently labeled ddNTPs, different lengths of extended primers immediately adjacent to the 5 '-end of the polymorphic site, and a PCR product template. The extension product is detected by an ABI sequencer, and based on this, the SNP site corresponding to the extension product and the type of the incorporated base can be determined, and finally the genotype of the sample can be determined. This technique is generally suitable for analyzing 10-30 SNP loci simultaneously. Direct sequencing is the most accurate of all SNP detection techniques and the gold standard for SNP analysis. Sequencing peaks of different types of sites can be obtained by directly amplifying and sequencing the fragments to be detected. This technology determines the genotype of the site by direct sequencing, which has the highest accuracy, but the cost is also high. If the SNP site to be detected is located in a sequencing unit, the cost of typing a single site can be significantly reduced, and this method is suitable for projects with small sample size or high accuracy requirements.
Real Experimental Research Hundreds of Detection Experiments 6 Experimental Platforms