RNA fluorescence in situ hybridization
A nucleic acid probe is a tool for detecting a specific nucleic acid molecule at the molecular level. For better qualitative, semi-quantitative or relative positioning analysis, specific nucleotides can be labeled with a reporter group (such as biotin or fluorescein). Such labeling can be visualized by immunochemical reaction or directly by fluorescence detection, so that the nucleic acid to be detected (such as mRNA, lncRNA, circRNA, miRNA) can be analyzed under the mirror. This method is relatively simple and has a wide range of applicability, so it is widely used in the field of biology.
Technical principle
RNA-FISH is an experimental method for analyzing a target nucleic acid in a cell or tissue section to be detected. This method uses homologous complementary nucleic acid probes to form hybrids with the target nucleic acid through a process of denaturation-annealing-renaturation. One of the nucleotides in the nucleic acid probe is labeled with a specific reporter molecule, such as biotin or fluorescein. Through immunochemical reaction or fluorescence detection system, using the role of reporter molecules and fluorescein labeled avidin, can be detected nucleic acids (mRNA, lncRNA, circRNA, miRNA) qualitative, semi-quantitative or relative positioning analysis. The principle of this method is clear, easy to operate, and can provide accurate and attractive experimental results.
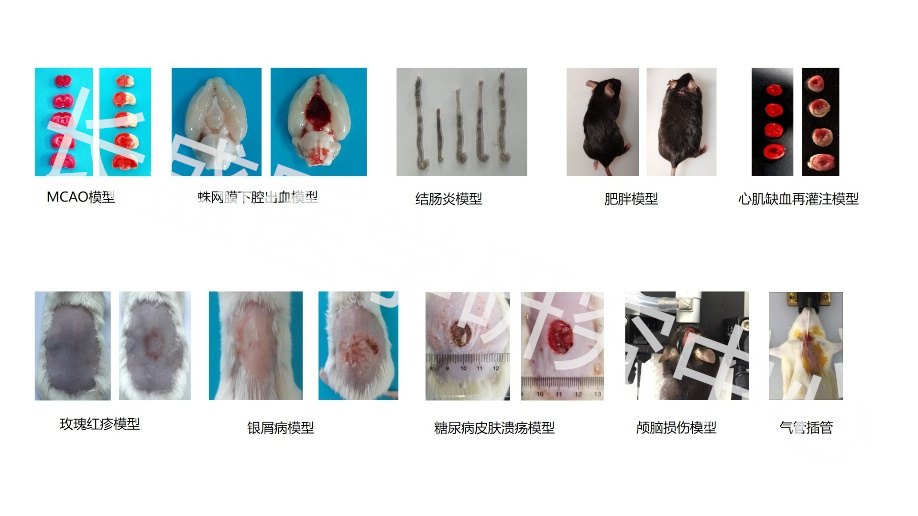
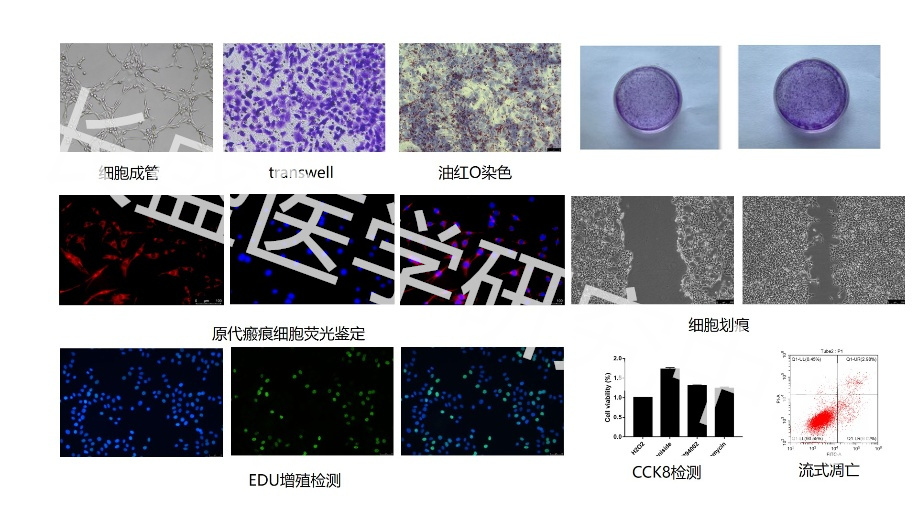
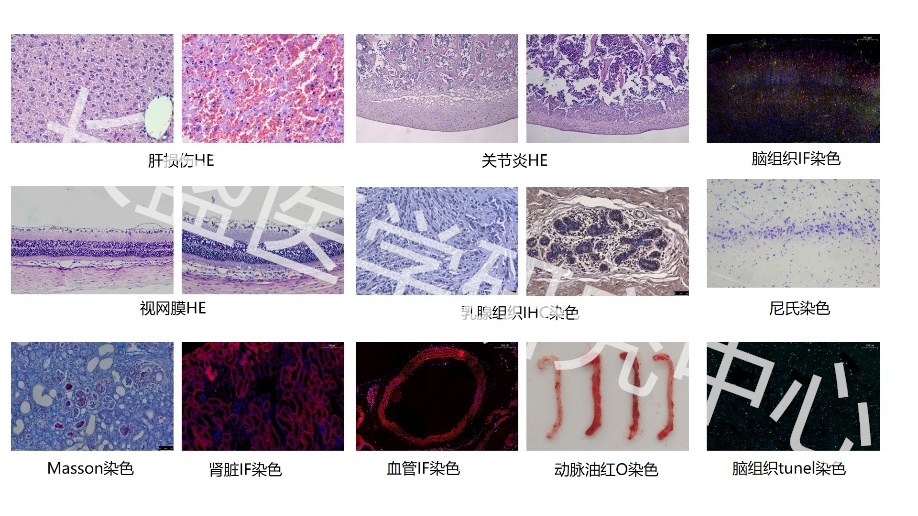
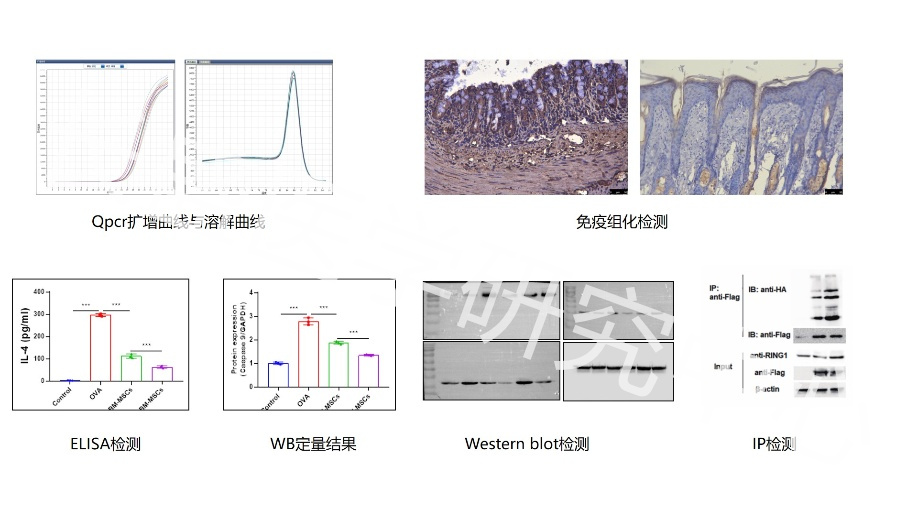
Real Experimental Research Hundreds of Detection Experiments 6 Experimental Platforms